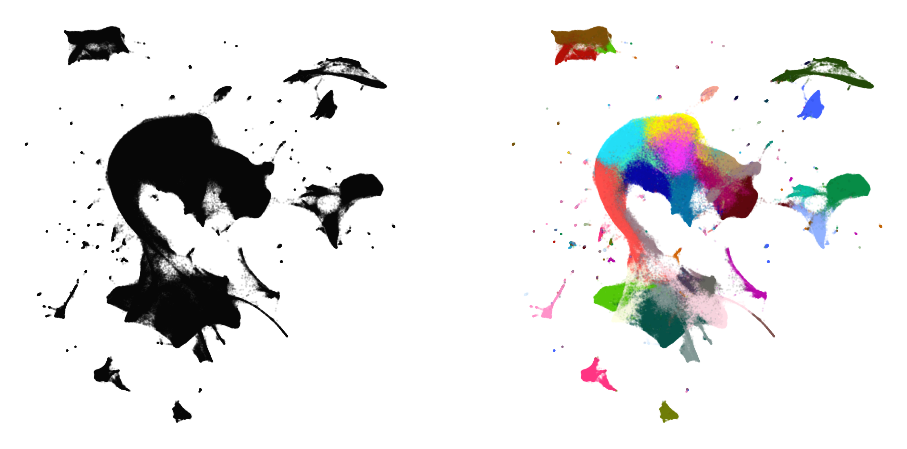Embedding large data sets
Embedding large data sets typically requires more care. Using various tricks described in preserving_global_structure can become quite slow to run. Instead, we can take a smaller, manageable sample of our data set, obtain a good visualization of that. Then, we can add the remaining points to the embedding and use that as our initialization.
Remember that the initialization largely affects the structure of the embedding. This way, our initialization provides the global structure for the embedding, and the subsequent optimization can focus on preserving local strucutre.
import gzip
import pickle
import numpy as np
import openTSNE
from examples import utils
import matplotlib.pyplot as plt
%matplotlib inline
Load data
The preprocessed data set can be downloaded from http://file.biolab.si/opentsne/benchmark/10x_mouse_zheng.pkl.gz.
%%time
with gzip.open("data/10x_mouse_zheng.pkl.gz", "rb") as f:
data = pickle.load(f)
x = data["pca_50"]
y = data["CellType1"]
CPU times: user 5.88 s, sys: 2.54 s, total: 8.42 s
Wall time: 8.42 s
print("Data set contains %d samples with %d features" % x.shape)
Data set contains 1306127 samples with 50 features
def plot(x, y, **kwargs):
fig, ax = plt.subplots(ncols=2, figsize=(16, 8))
alpha = kwargs.pop("alpha", 0.1)
utils.plot(
x,
np.zeros_like(y),
ax=ax[0],
colors={0: "k"},
alpha=alpha,
draw_legend=False,
**kwargs,
)
utils.plot(
x,
y,
ax=ax[1],
colors=utils.MOUSE_10X_COLORS,
alpha=alpha,
draw_legend=False,
**kwargs,
)
def rotate(degrees):
phi = degrees * np.pi / 180
return np.array([
[np.cos(phi), -np.sin(phi)],
[np.sin(phi), np.cos(phi)],
])
plot(x, y)
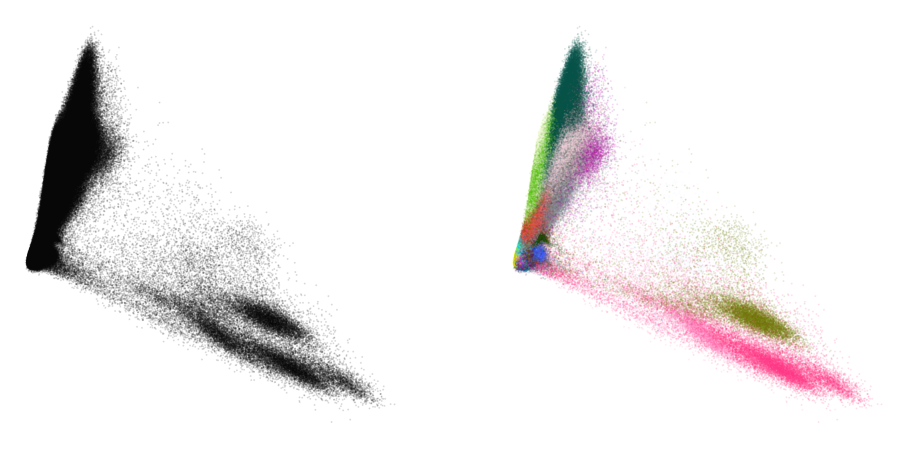
We’ll also precompute the full affinities, since we’ll be needing it in several places throughout the notebook, and can take a long time to run.
%%time
aff50 = openTSNE.affinity.PerplexityBasedNN(
x,
perplexity=50,
n_jobs=32,
random_state=0,
)
CPU times: user 52min 31s, sys: 1min 21s, total: 53min 53s
Wall time: 8min 2s
%%time
aff500 = openTSNE.affinity.PerplexityBasedNN(
x,
perplexity=500,
n_jobs=32,
random_state=0,
)
CPU times: user 5h 36min 15s, sys: 8min 53s, total: 5h 45min 8s
Wall time: 39min 40s
Standard t-SNE
First, let’s see what standard t-SNE does.
# Because we're given the data representation as the top 50 principal components
# we can just use the top 2 components as the initilization. There is no sense in
# calculating PCA on a PCA representation
init = openTSNE.initialization.rescale(x[:, :2])
%%time
embedding_standard = openTSNE.TSNE(
n_jobs=32,
verbose=True,
).fit(affinities=aff50, initialization=init)
--------------------------------------------------------------------------------
TSNE(early_exaggeration=12, n_jobs=32, verbose=True)
--------------------------------------------------------------------------------
===> Running optimization with exaggeration=12.00, lr=108843.92 for 250 iterations...
Iteration 50, KL divergence 8.0820, 50 iterations in 49.5503 sec
Iteration 100, KL divergence 7.8989, 50 iterations in 52.9387 sec
Iteration 150, KL divergence 7.8314, 50 iterations in 52.7589 sec
Iteration 200, KL divergence 7.8026, 50 iterations in 53.2414 sec
Iteration 250, KL divergence 7.7870, 50 iterations in 52.9956 sec
--> Time elapsed: 261.49 seconds
===> Running optimization with exaggeration=1.00, lr=1306127.00 for 500 iterations...
Iteration 50, KL divergence 5.7583, 50 iterations in 59.4852 sec
Iteration 100, KL divergence 5.4506, 50 iterations in 60.9192 sec
Iteration 150, KL divergence 5.2984, 50 iterations in 63.3280 sec
Iteration 200, KL divergence 5.2018, 50 iterations in 65.2476 sec
Iteration 250, KL divergence 5.1327, 50 iterations in 64.6718 sec
Iteration 300, KL divergence 5.0798, 50 iterations in 66.1166 sec
Iteration 350, KL divergence 5.0372, 50 iterations in 74.1704 sec
Iteration 400, KL divergence 5.0017, 50 iterations in 78.6565 sec
Iteration 450, KL divergence 4.9715, 50 iterations in 82.1395 sec
Iteration 500, KL divergence 4.9456, 50 iterations in 92.5772 sec
--> Time elapsed: 707.32 seconds
CPU times: user 3h 41min 29s, sys: 10min 8s, total: 3h 51min 37s
Wall time: 16min 14s
plot(embedding_standard, y)
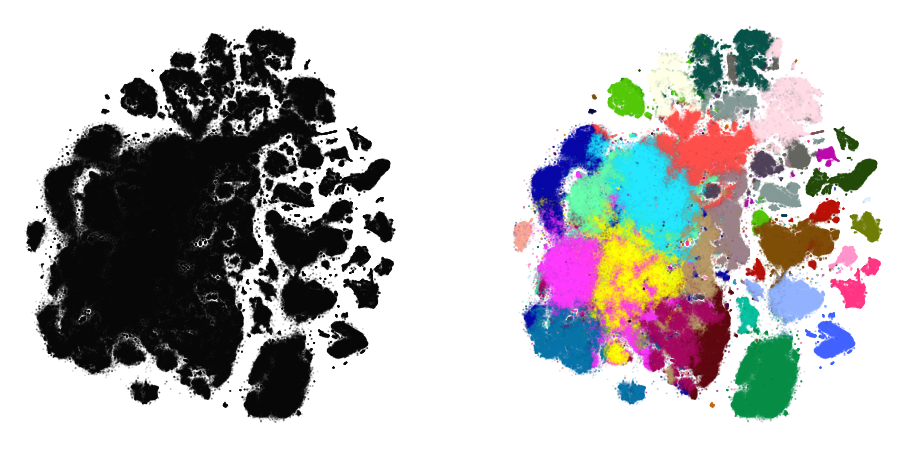
This doesn’t look too great. The cluster separation is quite poor and the visualization is visually not very appealing.
Using larger exaggeration
Exaggeration can be used in order to get better separation between clusters. Let’s see if that helps.
%%time
embedding_exag = openTSNE.TSNE(
exaggeration=4,
n_jobs=32,
verbose=True,
).fit(affinities=aff50, initialization=init)
--------------------------------------------------------------------------------
TSNE(early_exaggeration=12, exaggeration=4, n_jobs=32, verbose=True)
--------------------------------------------------------------------------------
===> Running optimization with exaggeration=12.00, lr=108843.92 for 250 iterations...
Iteration 50, KL divergence 8.0820, 50 iterations in 51.7397 sec
Iteration 100, KL divergence 7.8994, 50 iterations in 54.2033 sec
Iteration 150, KL divergence 7.8317, 50 iterations in 54.2117 sec
Iteration 200, KL divergence 7.8020, 50 iterations in 55.0897 sec
Iteration 50, KL divergence 6.9432, 50 iterations in 56.7796 sec
Iteration 100, KL divergence 6.8014, 50 iterations in 57.5679 sec
Iteration 150, KL divergence 6.7409, 50 iterations in 56.7147 sec
Iteration 200, KL divergence 6.7068, 50 iterations in 56.3419 sec
Iteration 250, KL divergence 6.6856, 50 iterations in 56.2568 sec
Iteration 300, KL divergence 6.6712, 50 iterations in 56.5754 sec
Iteration 350, KL divergence 6.6609, 50 iterations in 56.0285 sec
Iteration 400, KL divergence 6.6529, 50 iterations in 55.7784 sec
Iteration 450, KL divergence 6.6468, 50 iterations in 55.4943 sec
Iteration 500, KL divergence 6.6425, 50 iterations in 55.6567 sec
--> Time elapsed: 563.20 seconds
CPU times: user 3h 51min 45s, sys: 9min 50s, total: 4h 1min 35s
Wall time: 13min 57s
plot(embedding_exag, y)

The separation has improved quite a bit, but many clusters are still intertwined with others.
Using a larger perplexity
%%time
embedding_aff500 = openTSNE.TSNE(
n_jobs=32,
verbose=True,
).fit(affinities=aff500, initialization=init)
--------------------------------------------------------------------------------
TSNE(early_exaggeration=12, n_jobs=32, verbose=True)
--------------------------------------------------------------------------------
===> Running optimization with exaggeration=12.00, lr=108843.92 for 250 iterations...
Iteration 50, KL divergence 5.9688, 50 iterations in 216.9035 sec
Iteration 100, KL divergence 5.9218, 50 iterations in 218.9351 sec
Iteration 150, KL divergence 5.8993, 50 iterations in 218.5032 sec
Iteration 200, KL divergence 5.8900, 50 iterations in 223.6785 sec
Iteration 250, KL divergence 5.8854, 50 iterations in 224.3249 sec
--> Time elapsed: 1102.35 seconds
===> Running optimization with exaggeration=1.00, lr=1306127.00 for 500 iterations...
Iteration 50, KL divergence 3.8664, 50 iterations in 237.2816 sec
Iteration 100, KL divergence 3.6473, 50 iterations in 226.6633 sec
Iteration 150, KL divergence 3.5520, 50 iterations in 235.5178 sec
Iteration 200, KL divergence 3.4979, 50 iterations in 231.3676 sec
Iteration 250, KL divergence 3.4621, 50 iterations in 240.6245 sec
Iteration 300, KL divergence 3.4373, 50 iterations in 255.3066 sec
Iteration 350, KL divergence 3.4183, 50 iterations in 241.8503 sec
Iteration 400, KL divergence 3.4035, 50 iterations in 248.7666 sec
Iteration 450, KL divergence 3.3922, 50 iterations in 251.1434 sec
Iteration 500, KL divergence 3.3823, 50 iterations in 272.1862 sec
--> Time elapsed: 2440.72 seconds
CPU times: user 1d 2h 32min 21s, sys: 10min 19s, total: 1d 2h 42min 40s
Wall time: 59min 32s
plot(embedding_aff500, y)
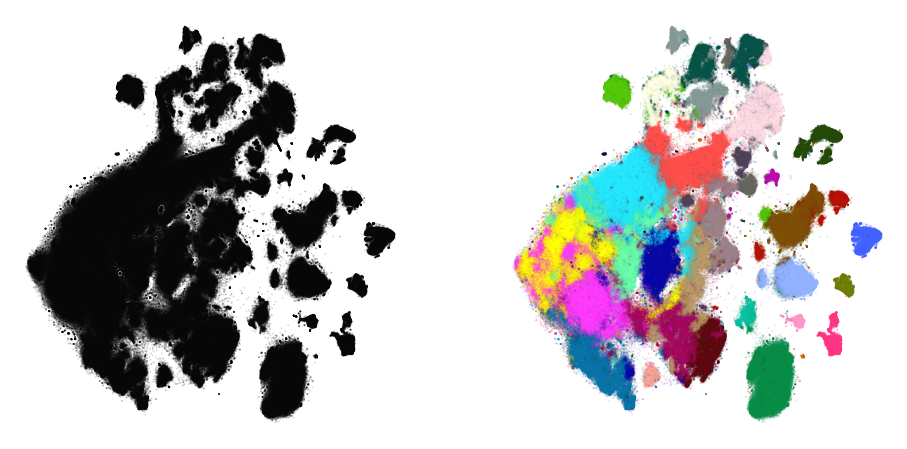
… with higher exaggeration
%%time
embedding_aff500_exag4 = openTSNE.TSNE(
exaggeration=4,
n_jobs=32,
verbose=True,
).fit(affinities=aff500, initialization=init)
--------------------------------------------------------------------------------
TSNE(early_exaggeration=12, exaggeration=4, n_jobs=32, verbose=True)
--------------------------------------------------------------------------------
===> Running optimization with exaggeration=12.00, lr=108843.92 for 250 iterations...
Iteration 50, KL divergence 5.9688, 50 iterations in 215.2427 sec
Iteration 100, KL divergence 5.9218, 50 iterations in 221.4924 sec
Iteration 150, KL divergence 5.8993, 50 iterations in 218.4626 sec
Iteration 200, KL divergence 5.8901, 50 iterations in 216.9501 sec
Iteration 250, KL divergence 5.8855, 50 iterations in 217.3419 sec
--> Time elapsed: 1089.49 seconds
===> Running optimization with exaggeration=4.00, lr=326531.75 for 500 iterations...
Iteration 50, KL divergence 5.0544, 50 iterations in 216.4466 sec
Iteration 100, KL divergence 4.9857, 50 iterations in 214.6171 sec
Iteration 150, KL divergence 4.9611, 50 iterations in 214.4493 sec
Iteration 200, KL divergence 4.9495, 50 iterations in 213.1030 sec
Iteration 250, KL divergence 4.9430, 50 iterations in 214.2775 sec
Iteration 300, KL divergence 4.9384, 50 iterations in 214.3649 sec
Iteration 350, KL divergence 4.9351, 50 iterations in 213.4982 sec
Iteration 400, KL divergence 4.9327, 50 iterations in 213.9199 sec
Iteration 450, KL divergence 4.9306, 50 iterations in 213.1906 sec
Iteration 500, KL divergence 4.9288, 50 iterations in 212.4853 sec
--> Time elapsed: 2140.36 seconds
CPU times: user 1d 1h 28min 18s, sys: 9min 56s, total: 1d 1h 38min 15s
Wall time: 54min 23s
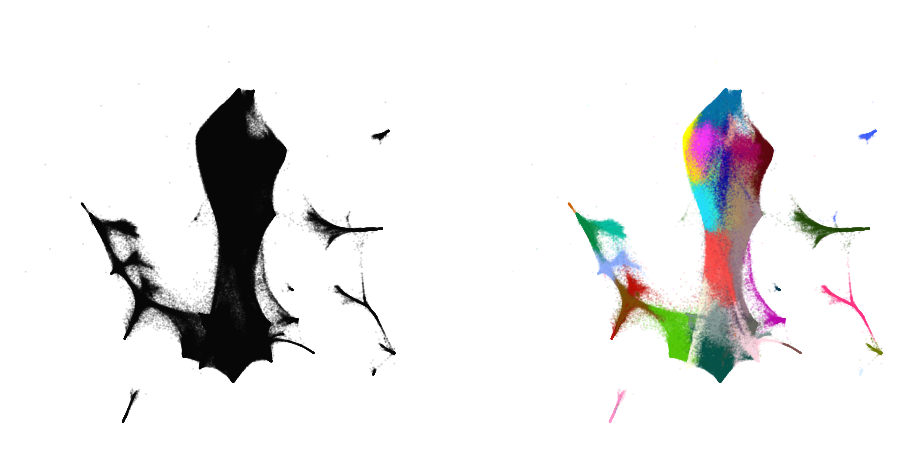
plot(embedding_aff500_exag4, y)
Initialize via downsampling
We now perform the sample-transform trick we described above.
Create train/test split
np.random.seed(0)
indices = np.random.permutation(list(range(x.shape[0])))
reverse = np.argsort(indices)
x_sample, x_rest = x[indices[:25000]], x[indices[25000:]]
y_sample, y_rest = y[indices[:25000]], y[indices[25000:]]
Create sample embedding
%%time
sample_affinities = openTSNE.affinity.PerplexityBasedNN(
x_sample,
perplexity=500,
n_jobs=32,
random_state=0,
verbose=True,
)
===> Finding 1500 nearest neighbors using Annoy approximate search using euclidean distance...
--> Time elapsed: 15.41 seconds
===> Calculating affinity matrix...
--> Time elapsed: 6.54 seconds
CPU times: user 3min 39s, sys: 4.97 s, total: 3min 44s
Wall time: 21.9 s
%time sample_init = openTSNE.initialization.pca(x_sample, random_state=42)
CPU times: user 5.35 s, sys: 657 ms, total: 6.01 s
Wall time: 171 ms
%time sample_embedding = openTSNE.TSNE(n_jobs=32, verbose=True).fit(affinities=sample_affinities, initialization=sample_init)
--------------------------------------------------------------------------------
TSNE(early_exaggeration=12, n_jobs=32, verbose=True)
--------------------------------------------------------------------------------
===> Running optimization with exaggeration=12.00, lr=2083.33 for 250 iterations...
Iteration 50, KL divergence 3.0347, 50 iterations in 4.0133 sec
Iteration 100, KL divergence 3.0733, 50 iterations in 3.9727 sec
Iteration 150, KL divergence 3.0691, 50 iterations in 4.0981 sec
Iteration 200, KL divergence 3.0690, 50 iterations in 3.9779 sec
Iteration 250, KL divergence 3.0690, 50 iterations in 3.9779 sec
--> Time elapsed: 20.04 seconds
===> Running optimization with exaggeration=1.00, lr=25000.00 for 500 iterations...
Iteration 50, KL divergence 1.1925, 50 iterations in 4.2847 sec
Iteration 100, KL divergence 1.1550, 50 iterations in 4.4657 sec
Iteration 150, KL divergence 1.1452, 50 iterations in 4.8737 sec
Iteration 200, KL divergence 1.1407, 50 iterations in 5.5286 sec
Iteration 250, KL divergence 1.1382, 50 iterations in 5.5675 sec
Iteration 300, KL divergence 1.1366, 50 iterations in 5.9546 sec
Iteration 350, KL divergence 1.1356, 50 iterations in 5.3339 sec
Iteration 400, KL divergence 1.1349, 50 iterations in 5.9090 sec
Iteration 450, KL divergence 1.1342, 50 iterations in 6.5074 sec
Iteration 500, KL divergence 1.1339, 50 iterations in 5.7942 sec
--> Time elapsed: 54.23 seconds
CPU times: user 34min 13s, sys: 2min 15s, total: 36min 28s
Wall time: 1min 14s
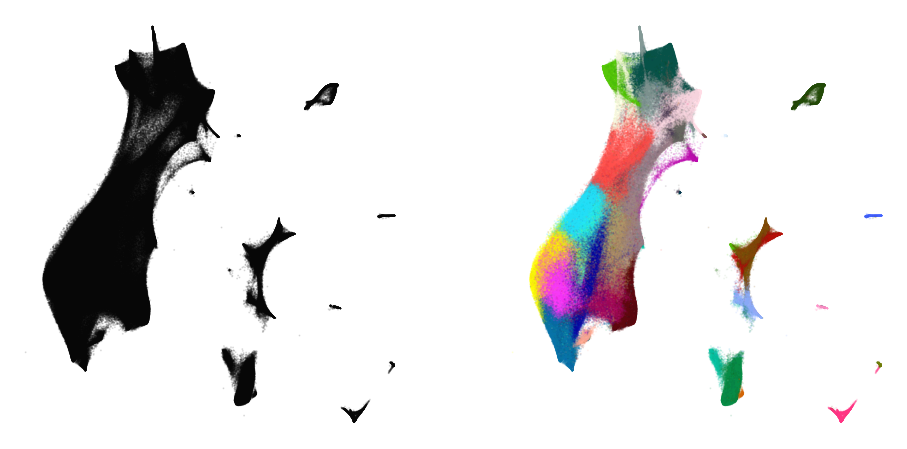
plot(sample_embedding, y[indices[:25000]], alpha=0.5)
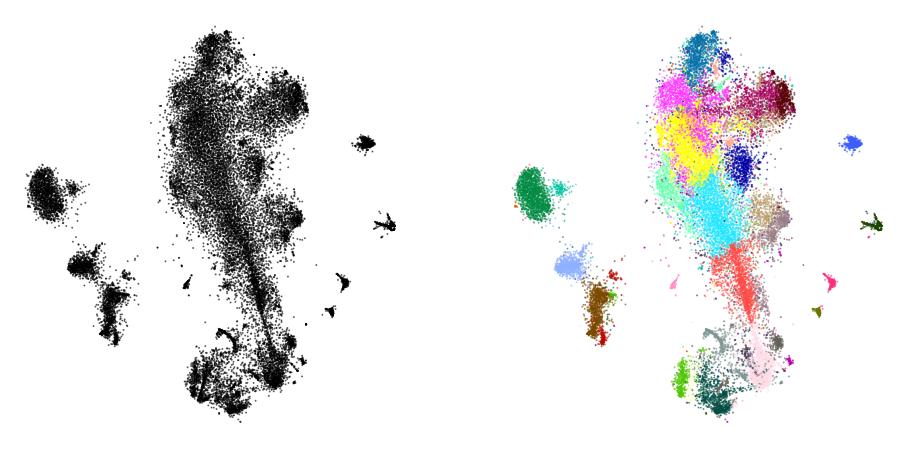
Learn the full embedding
%time rest_init = sample_embedding.prepare_partial(x_rest, k=1, perplexity=1/3)
===> Finding 1 nearest neighbors in existing embedding using Annoy approximate search...
--> Time elapsed: 174.82 seconds
===> Calculating affinity matrix...
--> Time elapsed: 0.67 seconds
CPU times: user 4min 18s, sys: 32.3 s, total: 4min 51s
Wall time: 2min 55s
init_full = np.vstack((sample_embedding, rest_init))[reverse]
plot(init_full, y)
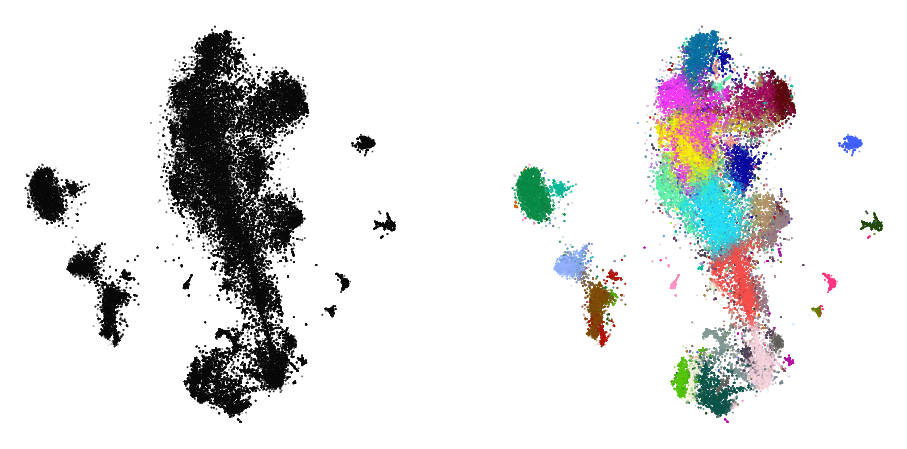
init_full = init_full / (np.std(init_full[:, 0]) * 10000)
np.std(init_full, axis=0)
array([1.00000000e-04, 1.06471542e-04])
embedding = openTSNE.TSNEEmbedding(
init_full,
aff50,
n_jobs=32,
verbose=True,
random_state=42,
)
%time embedding1 = embedding.optimize(n_iter=500, exaggeration=12)
===> Running optimization with exaggeration=12.00, lr=108843.92 for 500 iterations...
Iteration 50, KL divergence 7.9326, 50 iterations in 48.9843 sec
Iteration 100, KL divergence 7.8311, 50 iterations in 51.0335 sec
Iteration 150, KL divergence 7.7772, 50 iterations in 51.2970 sec
Iteration 200, KL divergence 7.7543, 50 iterations in 51.3436 sec
Iteration 250, KL divergence 7.7423, 50 iterations in 51.4951 sec
Iteration 300, KL divergence 7.7350, 50 iterations in 51.2477 sec
Iteration 350, KL divergence 7.7299, 50 iterations in 51.5019 sec
Iteration 400, KL divergence 7.7263, 50 iterations in 51.7564 sec
Iteration 450, KL divergence 7.7237, 50 iterations in 51.6552 sec
Iteration 500, KL divergence 7.7216, 50 iterations in 51.2847 sec
--> Time elapsed: 511.60 seconds
CPU times: user 2h 22min 35s, sys: 6min 28s, total: 2h 29min 4s
Wall time: 8min 34s
plot(embedding1, y)
%time embedding2 = embedding1.optimize(n_iter=250, exaggeration=4)
===> Running optimization with exaggeration=4.00, lr=326531.75 for 250 iterations...
Iteration 50, KL divergence 6.9325, 50 iterations in 55.0011 sec
Iteration 100, KL divergence 6.8182, 50 iterations in 54.9294 sec
Iteration 150, KL divergence 6.7678, 50 iterations in 54.4471 sec
Iteration 200, KL divergence 6.7398, 50 iterations in 54.4368 sec
Iteration 250, KL divergence 6.7219, 50 iterations in 53.8917 sec
--> Time elapsed: 272.71 seconds
CPU times: user 1h 13min 35s, sys: 3min 14s, total: 1h 16min 49s
Wall time: 4min 34s
plot(embedding2, y)
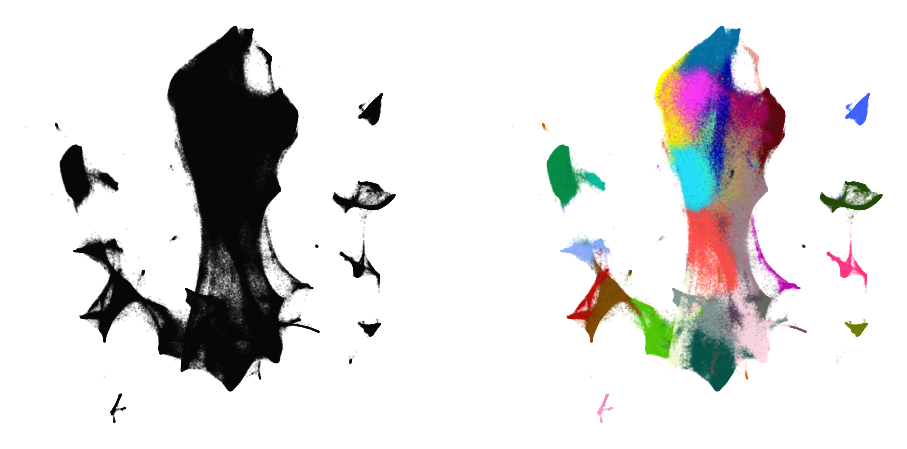
%time embedding3 = embedding2.optimize(n_iter=250, exaggeration=4)
===> Running optimization with exaggeration=4.00, lr=326531.75 for 250 iterations...
Iteration 50, KL divergence 6.7076, 50 iterations in 51.7026 sec
Iteration 100, KL divergence 6.6960, 50 iterations in 51.7740 sec
Iteration 150, KL divergence 6.6850, 50 iterations in 51.9709 sec
Iteration 200, KL divergence 6.6749, 50 iterations in 51.9325 sec
Iteration 250, KL divergence 6.6667, 50 iterations in 51.3449 sec
--> Time elapsed: 258.73 seconds
CPU times: user 1h 9min 44s, sys: 3min 14s, total: 1h 12min 58s
Wall time: 4min 20s
plot(embedding3, y)
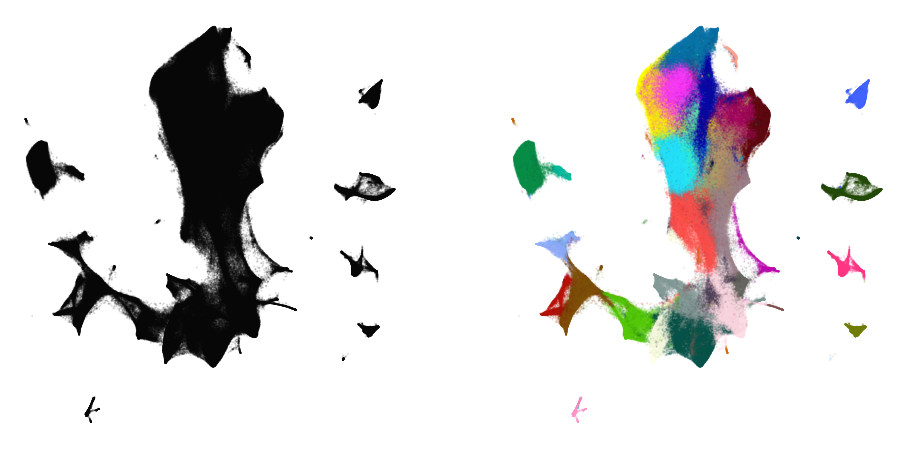
%time embedding4 = embedding3.optimize(n_iter=250, exaggeration=4)
===> Running optimization with exaggeration=4.00, lr=326531.75 for 250 iterations...
Iteration 50, KL divergence 6.6604, 50 iterations in 51.5464 sec
Iteration 100, KL divergence 6.6554, 50 iterations in 51.8160 sec
Iteration 150, KL divergence 6.6514, 50 iterations in 51.7537 sec
Iteration 200, KL divergence 6.6484, 50 iterations in 51.8397 sec
Iteration 250, KL divergence 6.6455, 50 iterations in 51.6008 sec
--> Time elapsed: 258.56 seconds
CPU times: user 1h 11min 20s, sys: 3min 14s, total: 1h 14min 35s
Wall time: 4min 20s
plot(embedding4, y)
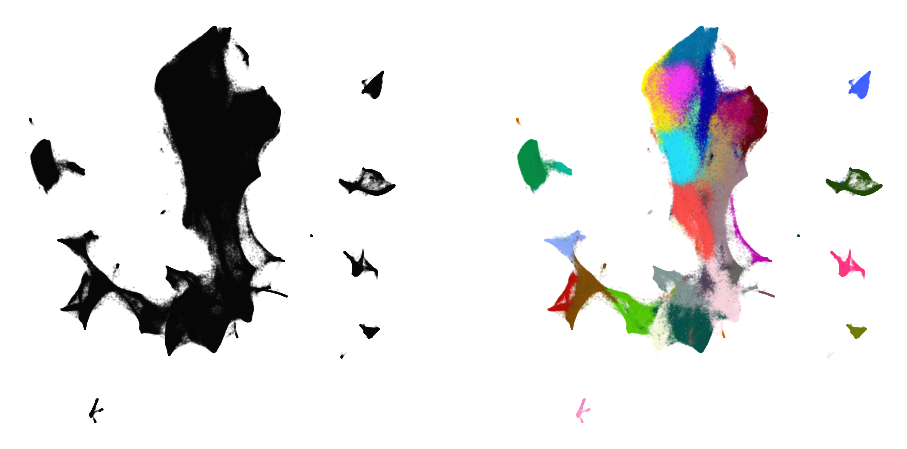
Comparison to UMAP
from umap import UMAP
/home/ppolicar/miniconda3/envs/tsne/lib/python3.10/site-packages/umap/distances.py:1063: NumbaDeprecationWarning: The 'nopython' keyword argument was not supplied to the 'numba.jit' decorator. The implicit default value for this argument is currently False, but it will be changed to True in Numba 0.59.0. See https://numba.readthedocs.io/en/stable/reference/deprecation.html#deprecation-of-object-mode-fall-back-behaviour-when-using-jit for details.
@numba.jit()
/home/ppolicar/miniconda3/envs/tsne/lib/python3.10/site-packages/umap/distances.py:1071: NumbaDeprecationWarning: The 'nopython' keyword argument was not supplied to the 'numba.jit' decorator. The implicit default value for this argument is currently False, but it will be changed to True in Numba 0.59.0. See https://numba.readthedocs.io/en/stable/reference/deprecation.html#deprecation-of-object-mode-fall-back-behaviour-when-using-jit for details.
@numba.jit()
/home/ppolicar/miniconda3/envs/tsne/lib/python3.10/site-packages/umap/distances.py:1086: NumbaDeprecationWarning: The 'nopython' keyword argument was not supplied to the 'numba.jit' decorator. The implicit default value for this argument is currently False, but it will be changed to True in Numba 0.59.0. See https://numba.readthedocs.io/en/stable/reference/deprecation.html#deprecation-of-object-mode-fall-back-behaviour-when-using-jit for details.
@numba.jit()
/home/ppolicar/miniconda3/envs/tsne/lib/python3.10/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
/home/ppolicar/miniconda3/envs/tsne/lib/python3.10/site-packages/umap/umap_.py:660: NumbaDeprecationWarning: The 'nopython' keyword argument was not supplied to the 'numba.jit' decorator. The implicit default value for this argument is currently False, but it will be changed to True in Numba 0.59.0. See https://numba.readthedocs.io/en/stable/reference/deprecation.html#deprecation-of-object-mode-fall-back-behaviour-when-using-jit for details.
@numba.jit()
umap = UMAP(n_neighbors=15, min_dist=0.1, random_state=1)
%time embedding_umap = umap.fit_transform(x)
OMP: Info #276: omp_set_nested routine deprecated, please use omp_set_max_active_levels instead.
CPU times: user 9h 39min 56s, sys: 1h 17min 49s, total: 10h 57min 45s
Wall time: 1h 21min 7s
plot(embedding_umap, y)